
9q34 & 16p13 chromosome duplications in autism
Introduction
The duplication or partial trisomy of chromosome 16p13 and chromosome 9q34 has been documented in the scientific literature (1,2). The duplication of chromosome 9 (trisomy 9p syndrome) is characterized by developmental delay, hypotonia, growth delay, facial and body malformations, and intellectual disability (ID). Additionally, other congenital defects in the heart, kidneys, skull, muscles, and nervous system have been identified (3). However, these defects vary according to which area of the chromosome is affected and by the length of the duplication (1,4). De novo 9q34 duplication presented with frontal bossing, deep-set eyes, facial anomalies (long and narrow), micrognathia, high nasal bridge, asymmetric ears, arachnodactyly, and other congenital malformations (5). The 9;15 translocation, 9q34 duplication, and 15q21q25 deletion resulted in ID, facial anomalies, hypotonia, and cardiac and renal malformations (6).
The partial trisomy of chromosome 16 duplication is characterized by ID, facial asymmetry, developmental delay, growth deficiency, and several congenital anomalies (7). Few cases of chromosome 16 duplication have been reported, as most individuals carrying the trisomy die during their childhood (2). The “critical” area at position 16p13.1–p13.3 was not affected in our patient (7,8). To our knowledge, region 16p13.13–p13.12 has previously been mentioned as a duplication, but no studies have been directed at this area (2). However, deletions of 16p13.13 delayed degradation of the Janus kinase 2 gene protein in B-cell lymphoma, and single nucleotide polymorphisms (SNPs) have been identified in cases of multiple sclerosis (9,10). Duplications of 16p13.13–p13.12 are yet to be understood.
We report the case of a 6-year-old male patient with non-consanguineous parents, carrying two duplications in chromosomes 9 and 16 and who had been diagnosed with autism spectrum disorder (ASD). Our patient presented with a mild form of trisomy 9 syndrome, but the characteristics associated with the duplication or partial duplication of chromosome 16 were—and remain—unclear.
Case presentation
Our patient was a 6-year-old boy with developmental delay. He was the first child of healthy, non-consanguineous Puerto Rican parents. His mother’s family history was remarkable for Jarcho-Levin syndrome (JLS). His mother had no previous abortions. He was born at 39 weeks of gestation to a 21-year-old mother. He weighed 5.2 lbs. and measured 48.26 cm. There were no complications during the pregnancy. At birth, he presented with macrocephaly and mildly dysmorphic facial features, but no body malformations were noted. We received him at 13 months of age at which time he evidenced macrocephaly and developmental delay. At 13 months, he could not sit by himself, which suggested psychomotor delay. He also presented with an abnormal curvature of the spine in the cervical area, with no other joint abnormalities. The physical examination revealed decreased muscle tone (hypotonia), dysmorphic features (micrognathia and macrocephaly), and a heart murmur.
Laboratory studies revealed high ammonia levels (148 ug/dL) and a lactate (18.8 mg/dL) to pyruvate (0.4 mg/dL) ratio of 47. Total and free carnitine levels were 68 and 50 umol/L (ratio =0.74) respectively. Therefore, he was treated for a metabolic disorder. We also ordered a karyotype analysis, which showed a normal 46, XY male karyotype with normal G-banding. Due to the child’s developmental delay, a magnetic resonance imaging was performed, the results of which showed that his ventricles, basal cisterns, and cerebral sulci all were prominent in size; no other abnormalities were seen. At 3 years of age, the child was clinically stable, but mental disability persisted. He was diagnosed with ASD after a psychological consultation.
Now 6 years of age, the patient is responsive and alert but is unable to verbalize. He also can walk normally and does not present body dysmorphic features, but mild hypotonia continues. His ears are symmetric but slightly larger than his parents’, measuring 5.5 cm; he has a head circumference of 54 cm. A microarray analysis was performed to determine whether the boy suffered from a genetic disease.
Results
A microarray analysis of peripheral blood sample showed approximately 982 kb duplication of the long arm of chromosome 9 (9q34.2 to 34.4) and approximately 1.6 Mb duplication of the short arm of chromosome 16 (16p13.13 to p13.12). Refer to Table 1.
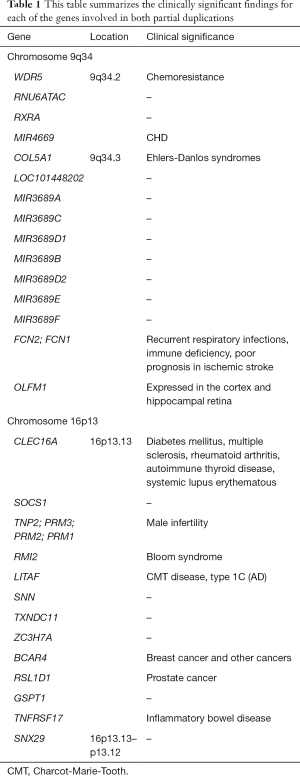
Full table
Chromosome 9 duplication
Partial duplications of genes WD repeat domain 5 (WDR5) and olfactomedin 1 (OLFM1) were identified, but the expressions of the proteins have not been analyzed, and therefore it is unclear whether their expression was affected by their partial duplication. In addition, complete duplications of genes RNA, U6atac small nuclear (U12-dependent splicing) (RNU6ATAC), retinoid X receptor alpha (RXRA), microRNA 4669 (miR4669), collagen type v alpha 1 chain (COL5A1), LOC101448202, microRNA 3689C (miR3689C), microRNA 3689D1 (miR3689D1), microRNA 3689B (miR3689B), microRNA3689D2 (miR 3689D2), microRNA 3689E (miR3689E), microRNA 3689F (miR3689F), ficolin 2 (FCN2), and ficolin 1 (FCN1) were also identified.
Chromosome 16 duplication
Partial duplications of C-type lectin domain containing 16A (CLEC16A) and sorting nexin 29 (SNX29) were identified, but their expressions were not submitted for analysis, and it is unclear whether their expressions have been affected by the partial duplication. In addition, complete duplications of genes suppressor of cytokine signaling 1 (SOCS1), transition protein 2 (TNP2), protamine 3 (PRM3), protamine 2 (PRM2), protamine 1 (PRM1), RecQ mediated genome instability 1 (RMI1), lipopolysaccharide induced TNF factor (LITAF), stannin (SNN), thioredoxin domain containing 11 (TXNDC11), zinc finger CCCH-type containing 7A (ZC3H7A), breast cancer anti-estrogen resistance 4 (BCAR4), ribosomal L1 domain containing 1 (RSL1D1), G1 to S phase transition 1 (GSPT1), and TNF receptor superfamily member 17 (TNFRSF17) were also identified.
Discussion
To our knowledge, none of the duplications discussed herein have been studied, or reported on in any medical or scientific publication. By analyzing the clinical relevance of each duplicated gene and observing the patient’s progress, we attempted to estimate and/or hypothesize the function of the gene or genes being analyzed. However, for genes RNU6ATAC, LOC101448202, SNN, TXNDC11, and ZC3H7A, no clinically relevant information was found.
WDR5 is expressed in greater quantities in the testes. Its overexpression has been linked to poor breast cancer prognosis and has been shown to promote both cancer-cell proliferation and chemo-resistance in bladder cancer (11,12). WDR5 has been shown to regulate bone growth in vivo (13).
RXRAs are located in the nuclei of various cells. RXRA has been shown to interact with vitamin D receptors to make a complex that interferes with RXRA chromatin localization and resists the inhibitory effects of 1α,25-dihydroxyvitamin D3. It is also being studied as a cancer therapy (14). Microduplication in 9q34.2–34.3 (RXRA’s location) was associated with congenital heart defects in 13 out of 25 patients (15).
Mutations in the COL5A1 gene are associated with Ehlers-Danlos syndromes and with life-threatening vascular abnormalities. However, only a small percentage of patients show vascular abnormalities (16). Our patient did not have any indications of a collagen disease.
Noncoding miRNAs have been described in the literature as regulators of several cellular mechanisms (17). Overexpressed miR569 has been identified in ovarian cancer (18). Additionally, Alzheimer’s disease, has been associated with the overexpression of hippocampal miR-34c, and mice lacking miR9-2 and miR9-3 have shown defects in mesencephalic structures (19,20). Our patient presented duplications in miR4669, miR3689A, miR3689C, miR3689D, miR3689B, miR3689D2, miR3689E, and miR3689F. To our knowledge, these ncRNAs have yet to be understood. Mutations in miR3689 are is a possible marker for B-cell acute lymphoblastic leukemia (21). We hypothesize that the duplication of one or more of these ncRNAs might be involved in brain development, as our patient was diagnosed with ASD. We also hypothesize that the overexpression of these genes was involved in the regulation of the other genes that were duplicated in this child, as he did not present the typical signs of 9q34 duplication syndrome.
FCN1 and FCN2 are involved in the immune response (22-24). FCN1 is expressed in granulocytes and monocytes and is released in secretory granules. It has been hypothesized that an increase of FCN1 might promote neutrophil adhesion, aggregation, and migration and consequently cause tissue damage. FCN2 levels decrease after ischemic stroke (25). There is a correlation between developing type 1 diabetes at an early age and SNP in FCN1, while SNP in FCN2 is associated with pulmonary disease (23,24,26). FCN2 has been shown to enhance phagocytosis, and low levels of this protein are related to immune deficiency and recurrent respiratory infections (22,24,27). Our patient did not present problems with his immune system.
OLFM1 is mainly expressed in the cortex and hippocampal retina, and is associated with neurogenesis, cell-cycle regulation, and tumorigenesis (28-30). In animal models, the overexpression of OLFM1 causes excessive migration of the neural tube, enlargement of the neural tube, and thickening of the optic tectum (31-33). The inhibition of OLFM1 causes ocular problems, such as reduced eye size and poor extension of the optic nerve (33). In mice, OLFM1 interacts with schizophrenia-1-gene protein (34). These findings together with the phenotype observed in our patient could suggest that OLFM1 might also be related to ASD development in humans. However, more studies should be done to substantiate this claim.
There have been several genome-wide association studies that have related CLEC16A to the development of several diseases, such as systemic lupus erythematosus, multiple sclerosis, type one diabetes, juvenile arthritis, alopecia, rheumatoid arthritis, primary adrenal insufficiency, and biliary cirrhosis (35-43). CLEC16A is important for autolysosome function in Purkinje cells, and homozygous mutations in this gene cause neurodegenerative disease that manifests as motor impairment (44). Mice lacking CLEC16A presented with low levels of B cells and immunodeficiency (45).
The SOCS1 protein interacts with IFN-alpha receptor 1 and the TNF-gamma receptor to inhibit STATs activation. SOCS1 can also attenuate Toll-like receptor-mediated modulation of T-cell response. Its overexpression might protect against multiple sclerosis and tumor development because of its JAK/STAT attenuation effects (46).
Transition nuclear protein two gene mutations have been associated with reduced and abnormal sperm and, consequently, infertility (in mice) (47). Another study identified SNPs in the TNP2 genes of infertile human males (48). PRM1 and PRM2 SNPs have been strongly associated with male infertility because of their essential function in sperm maturation (49). PRM3 has also been associated with sperm production (50). Duplications of these genes still need to be understood, but because of the role that these duplications have in chromatin condensation, it is possible that having increased levels of these proteins might lead to hyper-condensed chromatin.
Mutations in RecQ-mediated genome instability proteins 1 and 2 have been identified in a mild form of Bloom syndrome in a twin study (51). Xu et al. [2008], also associated RMIs with Bloom syndrome.
LITAF is a 161-amino acid protein that is associated with the highly hereditable Charcot-Marie-Tooth (CMT) type C disease (52,53). Our patient did not present any psychomotor delay during his first years of life.
Genetic abnormalities in BCAR4 have been implicated in breast cancer. The prognosis of patients with BCAR4 is worse when BCAR4 levels are highly elevated compared to those with lower levels of BCAR4 (54). High BCAR expression is also associated with poor prognosis in prostate cancer (55,56).
Li et al. [2016] associated the overexpression of RSL1D1 with poor prognosis in prostate cancer and postulated it as a possible biomarker of the disease.
Tumor necrosis factor superfamily 17 is involved in immune responses and B-cell maturation. This gene has been posited as being a marker for inflammatory bowel disease (57).
The grandmother of our patient had had two spontaneous abortions; both fetuses had been diagnosed with JLS. This syndrome is associated with the MESP2 gene, an abnormality in 9q34 (58). However, while our patient did not present a duplication in MESP2, this data could suggest that the family carries an anomaly in chromosome 9. A microarray analysis of gene expression should be performed on the mother to determine if she carries a mutation in chromosome 9. Finally, the phenotype presented by our patient could be associated with genetic factors, epigenetic changes, environmental influences, or a combination of these. The duplication in chromosome 16 might have a protective effect on the duplication in chromosome 9 or vice versa. Duplications in chromosome 16 are commonly lethal mutations, but patients that survive their presence have overlapping phenotypical features that are similar to those found in trisomy 9 syndrome. However, due to the lethality of trisomy 16 a well-established diagnosis has yet to be developed.
Acknowledgments
We would like to acknowledge the staff at San Jorge Hospital, medical office 408, and Ponce Health Sciences University for making it possible to write this clinical report. In addition, our thanks go to Bob Ritchie of the Publications Office of Ponce Health Sciences University-Ponce Research Institute.
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/acr.2020.03.07). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from the parents of the patient for the publication of this case report.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Allderdice PW, Eales B, Onyett H, et al. Duplication 9q34 Syndrome. Am J Hum Genet 1983;35:1005-19. [PubMed]
- Digilio MC, Bernardini L, Capalbo A, et al. 16p subtelomeric duplication: a clinically recognizable syndrome. Eur J Hum Genet 2009;17:1135-40. [Crossref] [PubMed]
- Kannan TP, Hemlatha S, Ankathil R, et al. Clinical manifestations in trisomy 9. Indian J Pediatr 2009;76:745-6. [Crossref] [PubMed]
- Jones KL, Jones MC. Duplication 9p Syndrome: (Trisomy 9p syndrome). Smith’s Recognizable Patterns of Human Malformation. Philadelphia: Elsevier Saunders, 2013.
- Youngs EL, McCord T, Hellings JA, et al. An 18-year follow-up report on an infant with a duplication of 9q34. Am J Med Genet A 2010;152A:230-3. [Crossref] [PubMed]
- Papadopoulou E, Sismani C, Christodoulou C, et al. Phenotype-genotype correlation of a patient with a “balanced” translocation 9;15 and cryptic 9q34 duplication and 15q21q25 deletion. Am J Med Genet A 2010;152A:1515-22. [PubMed]
- Ciaccio C, Tucci A, Scuvera G, et al. 16p13 microduplication without CREBBP involvement: moving toward a phenotype delineation. Eur J Med Genet 2017;60:159-62. [Crossref] [PubMed]
- Mohamed AM, Kamel A, Mahmoud W, et al. Intellectual disability secondary to a 16p13 duplication in a 1;16 translocation. Extended phenotype in a four-generation family. Am J Med Genet A 2015;167A:128-36. [Crossref] [PubMed]
- Melzner I, Weniger MA, Bucur AJ, et al. Biallelic deletion within 16p13.13 including SOCS-1 in Karpas1106P mediastinal B-cell lymphoma line is associated with delayed degradation of JAK2 protein. Int J Cancer 2006;118:1941-4. [Crossref] [PubMed]
- Zuvich RL, Bush WS, McCauley JL, et al. Interrogating the complex role of chromosome 16p13.13 in multiple sclerosis susceptibility: independent genetic signals in the CIITA-CLEC16A-SOCS1 gene complex. Hum Mol Genet 2011;20:3517-24. [Crossref] [PubMed]
- Dai X, Guo W, Zhan C, et al. WDR5 expression is prognostic of breast cancer outcome. PLoS One 2015;10:e0124964. [Crossref] [PubMed]
- Chen X, Xie W, Gu P, et al. Upregulated WDR5 promotes proliferation, self-renewal and chemoresistance in bladder cancer via mediating H3K4 trimethylation. Sci Rep 2015;5:8293. [Crossref] [PubMed]
- Gori F, Friedman L, Demay MB. Wdr5, a novel WD repeat protein, regulates osteoblast and chondrocyte differentiation in vivo. J Musculoskelet Neuronal Interact 2005;5:338-9. [PubMed]
- Jusu S, Presley JF, Kremer R. Phosphorylation of human retinoid X receptor α at serine 260 impairs its subcellular localization, receptor interaction, nuclear mobility, and 1 α,25-dihydroxyvitamin D3-dependent DNA binding in Ras-transformed keratinocytes. J Biol Chem 2017;292:1490-9. [Crossref] [PubMed]
- Amarillo IE, O’Connor S, Lee CK, et al. De novo 9q gain in an infant with tetralogy of Fallot with absent pulmonary valve: Patient report and review of congenital heart disease in 9q duplication syndrome. Am J Med Genet A 2015;167A:2966-74. [Crossref] [PubMed]
- Monroe GR, Harakalova M, van der Crabben SN, et al. Familial Ehlers-Danlos Syndrome with lethal arterial events caused by a mutation in COL5A1. Am J Med Genet A 2015;167:1196-203. [Crossref] [PubMed]
- Tuna M, Machado AS, Calin GA. Genetic and epigenetic alterations of MicroRNAs and implications for human cancers and other diseases. Genes Chromosomes Cancer 2016;55:193-214. [Crossref] [PubMed]
- Chaluvally-Raghavan P, Zhang F, Pradeep S, et al. Copy number gain of has-miR-569 at 3q26.2 leads to loss of TP531NP1 and aggressiveness of epithelial cancers. Cancer Cell 2014;26:863-79. [Crossref] [PubMed]
- Wang X, Liu P, Zhu H, et al. miR-34a, a microRNA up-regulated in a double transgenic mouse model of Alzheimer’s disease, inhibits bcl2 translation. Brain Res Bull 2009;80:268-73. [Crossref] [PubMed]
- Shibata M, Nakao H, Kiyonari H, et al. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci 2011;31:3407-22. [Crossref] [PubMed]
- Gutierrez-Camino A, Martin-Guerrero I, Dolzan V, et al. Involvement of SNPs in miR-3117 and miR-3689d2 in childhood acute lymphoblastic leukemia risk. Oncotarget 2018;9:22907-14. [Crossref] [PubMed]
- Faik I, Oyedeji SI, Idris Z, et al. Ficolin-2 levels and genetic polymorphism of FCN2 in malaria. Hum Immunol 2011;72:74-9. [Crossref] [PubMed]
- Xu DD, Wang C, Jiang F, et al. Association of the FCN2 gene single nucleotide polymorphisms with susceptibility to pulmonary tuberculosis. PLoS One 2015;10:e0138356. [Crossref] [PubMed]
- Metzger ML, Michelfelder I, Goldacker S, et al. Low ficolin-2 levels in common variable immunodeficiency patients with bronchiectasis. Clin Exp Immunol 2015;179:256-64. [Crossref] [PubMed]
- Zangari R, Zanier ER, Torgano G, et al. Early ficolin-1 a sensitive prognostic marker for functional outcome in ischemic stroke. J Neuroinflammation 2016;13:16. [Crossref] [PubMed]
- Anjosa ZP, Santos MM, Rodrigues NJ, et al. Polymorphism in ficolin-1 (FCN1) gene is associated with an earlier onset of type 1 diabetes mellitus in children and adolescents from northeast Brazil. J Genet 2016;95:1031-4. [Crossref] [PubMed]
- Matsushita M, Endo Y, Hamasaki N, et al. Activation of the lectin complement pathway by ficolins. Int Immunopharmacol 2001;1:359-63. [Crossref] [PubMed]
- Stone EM, Fingert JH, Alward WL, et al. Identification of a gene that causes primary open angle glaucoma. Science 1997;275:668-70. [Crossref] [PubMed]
- Funayama T, Mashima Y, Ohtake Y, et al. SNPs and interaction analyses of noelin 2, myocilin, and optineurin genes in Japanese patients with open-angle glaucoma. Invest Ophthalmol Vis Sci 2006;47:5368-75. [Crossref] [PubMed]
- Tomarev SI, Nakaya N. Olfactomedin domain-containing proteins: possible mechanisms of action and functions in normal development and pathology. Mol Neurobiol 2009;40:122-38. [Crossref] [PubMed]
- Barembaum M, Moreno TA, LaBonne C, et al. Noelin-1 is a secreted glycoprotein involved in generation of the neural crest. Nat Cell Biol 2000;2:219-25. [Crossref] [PubMed]
- Moreno TA, Bronner-Fraser M. Noelins modulate the timing of neuronal differentiation during development. Dev Biol 2005;288:434-47. [Crossref] [PubMed]
- Nakaya N, Lee HS, Takada Y, et al. Zebrafish olfactomedin 1 regulates retinal axon elongation in vivo and is a modulator of Wnt signaling pathway. J Neurosci 2008;28:7900-10. [Crossref] [PubMed]
- Camargo LM, Collura V, Rain JC, et al. Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol Psychiatry 2007;12:74-86. [Crossref] [PubMed]
- Berge T, Leikfoss IS, Harbo HF. From Identification to Characterization of the Multiple Sclerosis Susceptibility Gene CLEC16A. Int J Mol Sci 2013;14:4476-97. [Crossref] [PubMed]
- Hakonarson H, Grant SF, Bradfield JP, et al. A genome-wide association study identifies KIAA0350 as type 1 diabetes gene. Nature 2007;448:591-4. [Crossref] [PubMed]
- Hirschfield GM, Xie G, Lu E, et al. Association of primary biliary cirrhosis with variants in the CLEC16A, SOCS1, SPIB, and SIAE immunomodulatory genes. Genes Immun 2012;13:328-35. [Crossref] [PubMed]
- Jagielska D, Redler S, Brockschmidt FF, et al. Follow-up study of the first genome-wide association scan in alopecia areata: IL13 and KIAA0350 as susceptibility loci supported with genome-wide significance. J Invest Dermatol 2012;132:2192-7. [Crossref] [PubMed]
- Skinningsrud B, Lie BA, Husebye ES, et al. A CLEC16A variant confers risk for juvenile idiopathic arthritis and anti-cyclic citrullinated peptide antibody negative rheumatoid arthritis. Ann Rheum Dis 2010;69:1471-4. [Crossref] [PubMed]
- Skinningsrud B, Husebye ES, Pearce SH, et al. Polymorphisms in CLEC16A and CIITA at 16p13 are associated with primary adrenal insufficiency. J Clin Endocrinol Metab 2008;93:3310-7. [Crossref] [PubMed]
- Leikfoss IS, Keshari PK, Gustavsen MW, et al. Multiple sclerosis risk allele in CLEC16A acts as an expression quantitative trait locus for CLEC16A and SOCS1 in CD4 T cells. PLoS One 2015;10:e0132957. [Crossref] [PubMed]
- Tam RC, Lee AL, Yang W, et al. Systemic lupus erythematous patients exhibit reduced expression of CLEC16A isoforms in peripheral leukocytes. Int J Mol Sci 2015;16:14428-40. [Crossref] [PubMed]
- Soleimanpour SA, Gupta A, Bakay M, et al. The diabetes susceptibility gene Clec16a regulates mitophagy. Cell 2014;157:1577-90. [Crossref] [PubMed]
- Redmann V, Lamb CA, Hwang S, et al. Clec16a is critical for autolysosome function and purkinje cell survival. Sci Rep 2016;6:23326. [Crossref] [PubMed]
- Li J, Jørgensen SF, Maggadottir SM, et al. Association of CLEC16A with human common variable immunodeficiency disorder and role in murine B cells. Nat Commun 2015;6:6804. [Crossref] [PubMed]
- Baker BJ, Akhtar LN, Benveniste EN. SOCS1 and SOCS3 in the control of CNS immunity. Trends Immunol 2009;30:392-400. [Crossref] [PubMed]
- Tseden K, Topaloglu O, Meinhardt A, et al. Premature translation of transition protein 2 mRNA causes sperm abnormalities and male infertility. Mol Reprod Dev 2007;74:273-9. [Crossref] [PubMed]
- Miyagawa Y, Nishimura H, Tsujimura A, et al. Single-nucleotide polymorphisms and mutation analyses of the TNP1 and TNP2 genes of fertile and infertile human male populations. J Androl 2005;26:779-86. [Crossref] [PubMed]
- Jiang W, Sun H, Zhang J, et al. Polymorphism in protamine 1 and protamine 2 predict the risk of male infertility: a meta-analysis. Sci Rep 2015;5:15300. [Crossref] [PubMed]
- Oliva R. Protamines and male infertility. Hum Reprod Update 2006;12:417-35. [Crossref] [PubMed]
- Hudson DF, Amor DJ, Boys A, et al. Loss of RMI2 increases genome instability and causes a Bloom-like syndrome. PLoS Genet 2016;12:e1006483. [Crossref] [PubMed]
- Myokai F, Takashiba S, Lebo R, et al. A novel lipopolysaccharide- induced transcription factor regulating tumor necrosis factor alpha gene expression: Molecular cloning, sequencing, characterization, and chromosomal assignment. Proc Natl Acad Sci U S A 1999;96:4518-23. [Crossref] [PubMed]
- Lacerda AF, Hartjes E, Brunetti CR. LITAF mutations associated with Charcot-Marie-Tooth disease 1C show mislocalization from the late endosome/lysosome to the mitochondria. PLoS One 2014;9:e103454. [Crossref] [PubMed]
- Gong J, Zhang H, He L, et al. Increased expression of long non-coding RNA BCAR4 is predictive of poor prognosis in patients with non-small cell lung cancer. Tohoku J Exp Med 2017;241:29-34. [Crossref] [PubMed]
- Godinho MF, Sieuwerts AM, Look MP, et al. Relevance of BCAR4 in tamoxifen resistance and tumour aggressiveness of human breast cancer. Br J Cancer 2010;103:1284-91. [Crossref] [PubMed]
- van Agthoven T, Dorssers LC, Lehmann U, et al. Breast cancer anti-estrogen resistance 4 (BCAR4) drives proliferation of IPH-926 lobular carcinoma cells. PLoS One 2015;10:e0136845. [Crossref] [PubMed]
- Chae SC, Yu J, Oh GJ, et al. Identification of single nucleotide polymorphisms in the TNFRSF17 gene and their association with gastrointestinal disorders. Mol Cells 2010;29:21-8. [Crossref] [PubMed]
- Cornier AS, Staehling-Hampton K, Delventhal KM, et al. Mutations in the MESP2 gene cause spondylothoracic dystosis/Jarcho-Levin syndrome. Am J Hum Genet 2008;82:1334-41. [Crossref] [PubMed]
Cite this article as: Carlo SE, Martinez-Baladejo MT, Santiago-Cornier A, Arciniegas-Medina N. 9q34 & 16p13 chromosome duplications in autism. AME Case Rep 2020;4:17.


